A Pop Quiz for Cells
Photo courtesy of Pacific Northwest National Laboratory
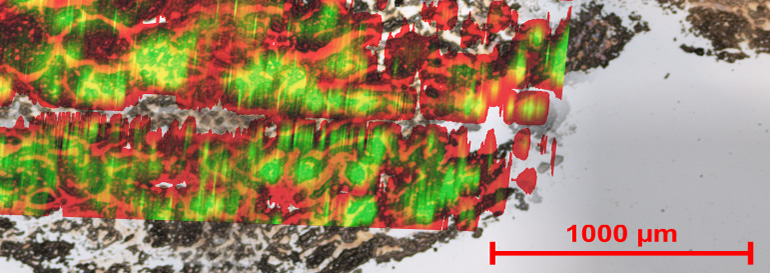
This sample shows an overlay of the optical and nanoDESI image of a human kidney tissue sample.
When trying to understand how cells respond to toxins, scientists want to do as little sample preparation as possible: preparing cells by immersing them in chemicals or drying them out can erase vital information. At PNNL, scientists proved that a new ionization technique they developed in 2009 can provide a fingerprint and locate proteins, amino acids, and other chemicals in cells that make up tissues or microbial communities using mass spectrometry. “The beauty of the technique is that it doesn’t require any sample preparation,” said Dr. Julia Laskin, who led the LDRD research project at PNNL. “Here, you just take your sample, slice it, and put it in front of the instrument for analysis.”
Why it Matters
Known as Nanospray Desorption Electrospray Iionization (nano-DESI), the technique efficiently allows scientists to determine which molecules reside in a precise spot on a sample. With this information, researchers can learn more about how diverse biological samples, such as tissues and microbes, respond to environmental factors. For example, biochemists can see how the marine microbe Shewanella oneidensis alters metals to remediatehazardous materials in the soil, or how S. oneidensis can change very soluble hexavalent uranium to less soluble form, limiting its movement in groundwater. Another opportunity lies with medical researchers learning how nicotine and other toxins affect brain cells.
“Great discoveries often require great tools,” said Dr. Louis Terminello, who leads PNNL’s Chemical Imaging Initiative. “The discoveries needed to solve today’s problems aren’t something that you’re going to get by eyeballing a sample.”
Methods
From the start, the team was convinced that the liquid bridge used in nano-DESI could be scaled down to analyze small areas on biological samples. With adjustments, the team scaled down the probe to analyze an area of 10 µm in diameter, about the size of a single red blood cell or mid-sized bacteria. “With this probe, we are getting down to individual cells,” said Laskin.
The team first analyzed rat brain tissues, which provide an outstanding test case for the technique, because they are very dense and yield high signals. The nano-DESI was able to draw up and analyze the molecules from different regions on the sample. Then, the team moved onto kidney tissues, and again analyzed micrometer-sized areas.
With each sample, the nanoDESI generated reams of mass spectra. Because existing software packages could not process the data, the work fell to team member Brandi Heath, who read the charts and determined the fatty acids, amino acids, lipids, and other molecules that resided at different locations on the tissue. “Compared to other online liquid extraction techniques, nano-DESI has about an order of magnitude better spatial resolution,” Laskin added. “It is comparable, spatially, to what laser-based techniques can give.”
What’s Next
The team is working on two efforts related to their work with nanoDESI, including understanding and visualizing the mass spectrometry data on the fly, and analyzing different microbial communities of DOE interest.
Publication
Laskin J, BS Heath, PJ Roach, L Cazares, and OJ Semmes. 2012. “Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry.” Analytical Chemistry 84(1):141-148. DOI: 10.1021/ac2021322
Acknowledgments
PNNL project team members include Julia Laskin, Brandi Heath, and Patrick Roach, PNNL; and Lisa Cazares and O. John Semmes at the Eastern Virginia Medical School provided the tissue samples.