Designing Drugs on Supercomputers
Researchers use Oak Ridge Leadership Computing Facility to accelerate drug discovery.
The Science
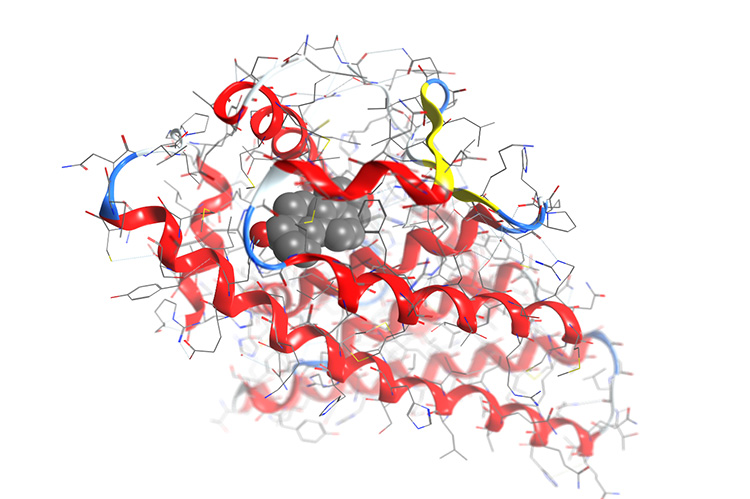
The Oak Ridge Leadership Computing Facility recently simulated tests of 2 million different drug compounds against a targeted receptor in less than two days.
The Impact
The average cost of developing and bringing one drug to market can range from a few hundred million dollars to more than a billion and taking from 10 to 15 years before patients get the medicines they need. The vast majority of prospective drug designs fail; therefore the industry wants them to “fail fast” and “fail cheap” in order to focus resources on the most promising candidates. Researchers are using the Oak Ridge Leadership Computing Facility to speed up the screening process while increasing the chance for developing a successful drug for a fraction of the cost.
Summary
Jerome Baudry, an assistant professor at the University of Tennessee (UT) and member of the Center for Molecular Biophysics at Oak Ridge National Laboratory (ORNL) and his team of computational biophysicists use supercomputers much like other scientists use microscopes. After making alterations to publicly licensed software from the Scripps Research Institute, they were able to create 3D biological simulations of compounds docking with receptors in the body and run it on one of the world’s fastest computers to screen millions of candidates in a few days. The simulations the team created are based upon the process by which molecular compounds function within the body. Pharmaceuticals work because they bind specifically to certain cellular receptors that play roles in health and disease; similar to the way a key fits a lock. When that key opens too many locks, however, side effects occur. Baudry and his collaborators want to be able to predict the specific binding of a drug to a receptor to avoid cross-reactivity. Knowing this behavior will help researchers generate drug candidates likely to survive clinical trials. Thanks to the efficient and massive computations possible using the Oak Ridge Leadership Computing Facility, Baudry and his collaborators can screen drug candidates against multiple receptors and the dynamic structural variations of those receptors. The ability to run simulations greatly reduces the sample size as poor drug candidates get eliminated and ultimately produces a more specifically binding, and therefore more efficient, drug.
Contact
Jerome Y. Baudry
baudryjy@ornl.gov
Funding
Calculations were performed using OLCF computing resources, supported by the Office of Science Advanced Scientific Computing Research (ASCR) program. The research is funded by a NIH Clinical Translational Science Award, which was awarded to Georgetown and Howard Universities and includes ORNL, Med/Star Health and the Washington D.C. Veterans Affairs Medical Center as key partners. Funding for the initial development work was provided by ORNL's Laboratory Directed Research and Development (LDRD) program.
Publications
Ellingson, S. R. and Baudry, J. High-throughput virtual molecular docking with AutoDockCloud. Concurrency Computat.: Pract. Exper. (2012). [DOI: 10.1002/cpe.2926]
Collignon, B., Schulz, R., Smith, J. C. and Baudry, J. Task-parallel message passing interface implementation of Autodock4 for docking of very large databases of compounds using high-performance super-computers. J. Comput. Chem., 32: 1202–1209. (2011) [DOI: 10.1002/jcc.21696]
Related Links
http://www.olcf.ornl.gov/2012/10/18/big-computing-cures-big-pharma-2/
Highlight Categories
Program: ASCR
Performer: University , DOE Laboratory , SC User Facilities , ASCR User Facilities , OLCF
Additional: Technology Impact , Collaborations , Non-DOE Interagency Collaboration